[under working]
This protocol is based on Rory Sayres‘ teaching.
Download and install MrVista
svn checkout https://white.stanford.edu/repos/vistasoft
If you already downloaded an older version, use
svn update https://white.stanford.edu/repos/vistasoft
Or you can download the zip file from
http://white.stanford.edu/software/
Setup data directory hierarchy
Inside a subject’s directory, say 13902, create directories
Raw
Raw/Pfiles
Raw/Anatomy/Inplane
Pay attention to the upper/lower cases.
Hint: To create nested folder, you may use -p option. e.g. mkdir abc/def/gh -p
Put inplane images (32 dicome files) into Raw/Anatomy/Inplane; put all retinotopy functional images into Raw/Pfiles. You should keep a track of which run is eccentricity or meridian (e.g. P18944 is ecc).
Functional analysis
- Start Matlab
- cd to MrVista root directory (/fs/quarry/cuixu/mrvista/) and add MrVista to MatLab path
addpath(genpath(pwd))
This step may take a long time (~1min) as there are a lot of folders. - cd to your subject’s directory (e.g. /fs/quarry/cuixu/retinotopy/data/13902/). You should stay in this directory when using mrvista.
- Initialize
mrInit2
Initialize window pops up (below).
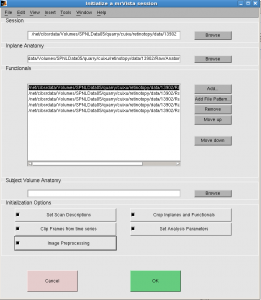
Select all 5 “Initialization Options” and click OK.
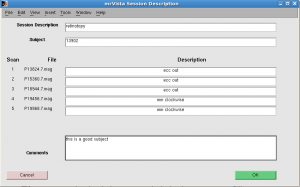
Session Description: retinotopy
Subject: <your subject’s name> e.g. 13902
For each run, fill either “ecc” or “mm”. You can add more info such as “ecc out” or “mm clockwise”. Click OK.
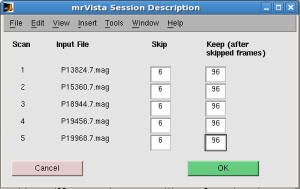
Skip: 6 scans. Keep 96 scans for all runs.
Crop Inplanes window:
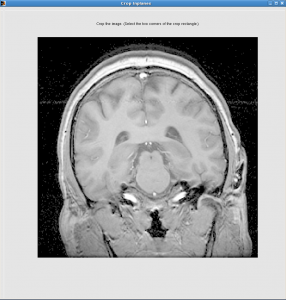
Select a slice where brain is the biggest
Click the upper-left point, and then lower-right point, the rectangle between will be kept. You should keep the skull but not much neck.
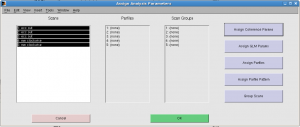
In “Assign analysis parameters”, select all scans, click “Assign coherence params” and input 8 as “Stimulus frequency cycles/scan”.
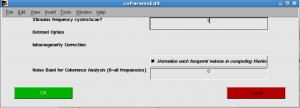
Click OK.
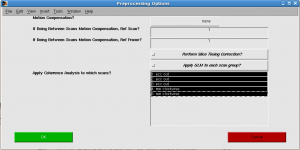
In “Preprocessing Options” window, select all scans and click OK. Progress will be shown. You may encounter warning “loadtSeries: unexpected number of tSeries frames in file.”. It’s a bug. (For now, you can set the nfrmaes to 96 by mrSESSION.functionals(1).totalFrames=96;)
- Run mrvista
mrvista
MrVista main window pops up.
Resize and click menu File | Save preferences to save preferences (e.g. window size and positoin).
You may also want to change 1st_Slice and #_slices to have different views (e.g. display multiple slices at once)
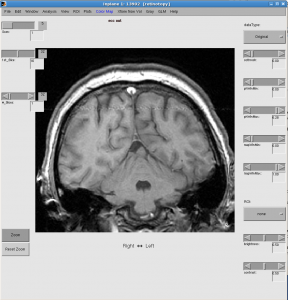
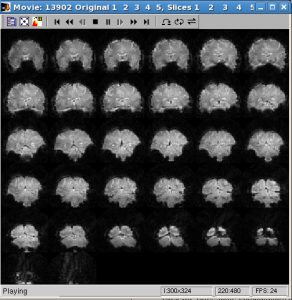
- Check data quality by showing movie
Click Menu Plots | tSeries Movie | Movie UI (set params)
Scans: 1:5 (the number of runs)
Slices: 1:32 (the number of slices in inplane)
Click OK. After some computation, movie will be shown. With movie you can check visually if subject’s move their head a lot within run.
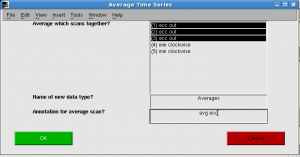
- Average all ecc scans, and mm scans.
click Analysis | Time Series | Average tSeries. “Average Time Series” window pops up. Select all ecc runs, and give annotation “avg ecc”. Progress bar shows.
Repeat for mm.
- Select dataType as “Averages” (previously it is “original”) on the upper-right corner of mrvista main interface.
- Do analysis on averaged scans.
click menu Analysis | Traveling Wave Analysis | compute corAnal | computer corAnal (all scans) - View coherence map: View | Coherence map. You see the back of brain (visual cortex) has higher coherence with stimuli (a lot of red). Coherence is kind of correlation regardless lag.
- View amplitude map: View | amplitude map. Amplitude is usually low so we need to change colorbar. Right click the color bar in the bottom and select context menu “Clip mode”. A window pops up. Input 0 4 to see differential colors for amplitude.
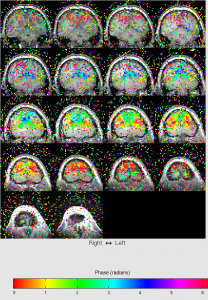
- View phase map: View | phase map. This is most important map.
For ecc, you should see that color changes from blue to red when moving from front to back of the head.
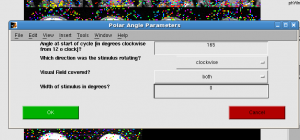
For mm, you first need to set retinotopy parameters. Click Edit | Data Type | Set retinotopy parameter | current scan. Start:165, clockwise, width 0. Click OK. You will find the color bar change to polar.
You should see the upper left of brain is redish, upper right of brain is yellowish.
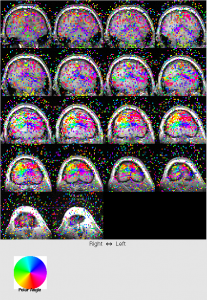
- ROI
First you may need to display only 1 slice to zoom ot.
Create ROI: ROI | Create | Create Rectangle ROI. Click two points.
Restrict ROI (to super-threshold voxels): ROI | restrict | restrict selected ROI - Plot ROI time series or FFT
Click Plots | Current scan | mean time series to view time series. You should see 8 cycles of up and downs.
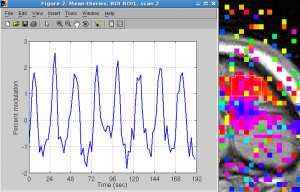
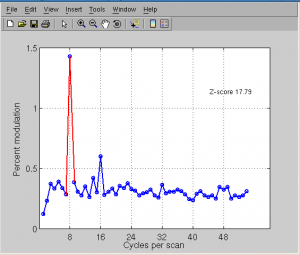
Click Plots | Current scan | mean fft to see frequency.
Anatomy processing
- Convert DICOM files to nifti file.
cd to the directory where the structural T1 image is. They are DICOM files (124 of them).
niftiFromDicom(pwd)
You will find a new file created (.nii.gz file). Rename it to t1_raw.nii.gz
Can’t make it work under scuttlebutt (mex file error), nor on rumor (stack error). But it works when I ssh scuttlebutt under my Mac and start as “ml7spm2 -jvm”. - Use mrViewer to identify if image directions are correct.
mrViewer t1_raw.nii.gz
Click “3-Axis View”
Select menu Coordinates > I|P|R
Select menu View > Label > Label directions
Check if directions LR, AP, IS are correct. If not, you will have to flip.
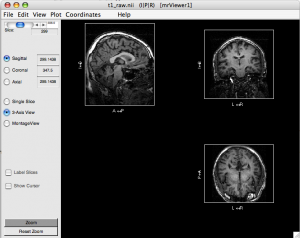
- Flip direction if necessary. If you find one direction is flipped (e.g. AP is flipped in the figure above), you need to correct it: (run these command on scuttlebutt, not Mac)
mr = mrLoad(‘t1_raw.nii.gz’);
mr.data = flipdim(mr.data, 2); % 2 means AP, 1 means LR, 3 means IS
mrSave(mr, ‘t1_raw_flipped.nii.gz’);
Confirm if flip take effect using mrViewer. - Reslice and ACPC the image
mrAnatAverageAcpcNifti({‘t1_raw_flipped.nii.gz’}, ‘t1.nii.gz’, [], [1,1,1]);
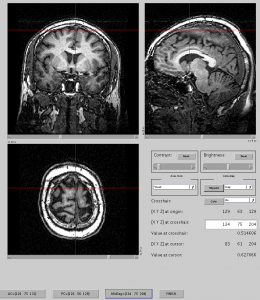
If you don’t know where is AC or PC, you need to refer my earlier post (how to find ACPC). - Now you should have t1.nii.gz which is already PCACed, isotropic (1mm). You can view this image with mrViewer.
- Run FIRST, initial segmentation. Note this command is run from shell, not within MatLab. This step takes ~2hr but you don’t have to wait to continue the next step.
run_first_all t1.nii.gz 4 t1_sgm - Run BET to strip skull
bet t1.nii.gz t1_bet.nii.gz -m -f .2
You may find you get t1_bet.img instead of t1_bet.nii.gz. If so, use mri_convert of freesurfer to convert image format. - Confirm skull stripping using mrViewer (or fslview).
Run mrViewer t1.nii.gz, then select menu File > Load > MR data map, select t1_bet.nii.gz file. You will see a red image. Adjust threshold (Threshold 1) slightly, you should see the skull-stripped image on top the original t1 image. Check if visual cortex is cut. If yes, you can re-run BET with different threshold.
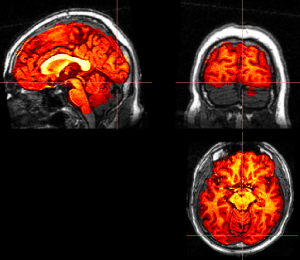
- Run FAST, segmentation on skull-stripped brain
fast t1_bet.nii.gz - Swap x dimension using fslswapdim
(After swapping I find left-right is not right in itkgray, so maybe should not swap?)
Can’t get it work in scuttlebutt. But we can achieve the same effect by flipdim in MatLab.
mr = mrLoad(‘t1_bet_seg.nii.gz’);
mr.data = flipdim(mr.data, 1); % 2 means AP, 1 means LR, 3 means IS
mrSave(mr, ‘t1_bet_seg_swap.nii.gz’); - Convert FIRST/FAST file to class file (takes ~1 min)
mrGrayConvertFirstToClass(‘t1_sgm_all_th4_first.nii.gz’,’t1_bet_seg_swap.nii.gz’,’t1_class.nii.gz’) - Use ITKGray for manual segmentation. ITKGray runs on linux. It has been already downloaded and installed at /quarry/cuixu/itkgray/itkGray_install/bin/itksnap
(You can download ITKGray from http://white.stanford.edu/software/downloads/itkGray/ITKGray_080128_fc4.tgz
unzip (tar -xzvf). Then run itksnap.)
Detailed ITKGray usage can be found at http://white.stanford.edu/newlm/index.php/ITKGray-Using
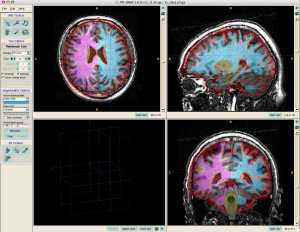
- In ITKGray, load original t1.nii.gz
File> Load Data> Greyscale Image (load t1.nii.gz)
Load segmentation data
File> Load Data> Segmentation Data (load t1_class.nii.gz) - Use Paint Brush tools to manually segment the brain. Overall label opacity is very useful – you need to change it constantly to identify where is gray/white matter.
Useful links: http://white.stanford.edu/newlm/index.php/Segementation-FSL-Pipeline
Related: stimulus parameters
| Parameters | ||||||
| sequence | spiral out | |||||
| implane slices | 32 | |||||
| TR | 2 | s | ||||
| Total number of frames | 108 | |||||
| N-extra | 4 | scan | 8 | s | ||
| After processing, the total number of images is 108, which corresponding to 216s. | ||||||
| 216 = 12 + 24*8 + 12 where 12s rest + 8 cycles of 24s task + 12s rest | ||||||
| During task and rest, subject perform the “red-press” task | ||||||
| Before the 1st rest, there is 8s count-down, which I believe corresponds to the 8s N-extra, which not included in the final processed images | ||||||
| mm | ||||||
| start angle | 165 | degree | ||||
| end angle | 135 | degree | ||||
| angle span | 45 | degree | ||||
| direction | clockwise | |||||
| 12s rest – 24s taskx8cycle – 12s rest | ||||||
| ecc | ||||||
| max visual angle | 28.5 | degree | ||||
| direction | out | |||||
| data location | ||||||
| p16_phunk\fmri\Tenforde_Adam\CON_WMS_Subjects | ||||||
| eyetracking data location | ||||||
| \\Cibsrdata\phunk\p16_phunk\fmri\Tenforde_Adam\CON_WMS_Subjects\eyetrackingdata | ||||||
| task location | ||||||
| \\Cibsrdata\vineyard\Ng_Derek\Ng_Derek\WMS_fMRI_Tasks | ||||||
| /fs/WILLIAMS/ | ||||||



Dear Cui,
I am newbie and also trying to do retinotopic mapping and, in your opinion, which approache is easier to track down? mrVista or FreeSurfer or BrainVoyager? Many thanks!
I never used BrainVoyager. As to MrVista and FreeSurfer, it’s hard to compare but I find it’s pretty difficult to use either without a real person to teach you.
AFNI now has a retinotopy pipeline – @RetinoProc. I think you need surfaces to use it; that should be fairly easy with Freesurfer’s recon-all (there are scripts for conversion between Freesurfer and AFNI).
Hope that helps.
Hi Xu Cui,
I am starting now with mrVista, until now I used BrainVoyager to do retinotopy.
I have a question about the functional files format that you put in Raw/Pfiles. Is it DICOM, as the anatomical files in Raw/Anatomy/Inplane? I tried with DICOM, but when I started mrVista the field Functionals was empty.
Thank you for your help and a happy new year!
Kind regards,
Sónia
Hi Xu Cui,
I have a question about the format of functional files that are in Raw/Pfiles. Is it DICOM as the anatomical files?
I am starting now with mrVista, usually I use BrainVoyager to do retinotopic analysis.
Thank you for your help.
Happy new year!
Kind regards,
Sónia (Portugal)
@Sónia Ferreira
Sónia, it has been a while since I used MrVista and I hate to give you inaccurate information. Please ask MrVista developers if you can.
Xu
Since we have no fMRI available and I want to understand retinotopy processing, I downloaded the data at https://white.stanford.edu/repos/vistadata/
which includes a folder called wmretinotopy, I assume it contains data related to retinotopy experiments. However, to be able to make use of them I need detailed explanation of what experiment exactly each file correspond to (need file descriptions relating data to detailed experiment explanation). I can’t find this anywhere. Can you help me?
Dear all,
this is Elena,
i m a brainvoyager user, however i really want to use mrvista to analyze prf.
i was able to run inplane (with tutorial from vistasoft) but when i want to see the gray i get error.
In general i really would like to learn vistasoft and mrvista but i feel a little lost, could you pls give me some help or tips? otherwise there is a forum/person where i can ask for help?
many many thx
elens