xjView’s official webpage moves to https://www.alivelearn.net/xjview/
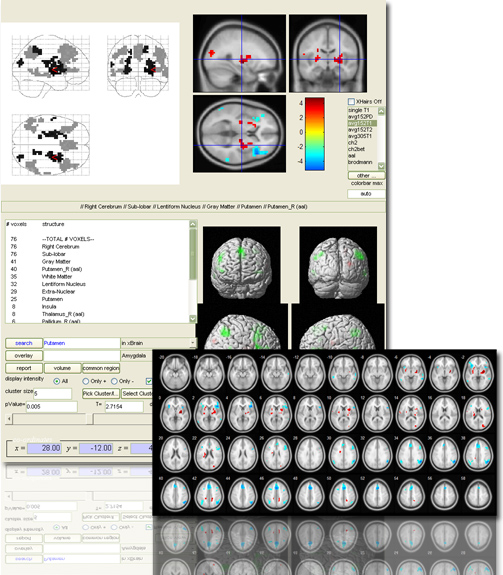
xjView is a MatLab program Jian and I wrote a few years ago to display SPM T-test images more easily. Main features include
- p-value slider: change p-value and display the supra-threshold activation instantly
- anatomical description: tell you if this is hippocampus or thalamus
- display more than one images together: so you can compare activations from different contrast
- single out an activation region and get statistics of “how many voxels are in what region”
xjView also has a powerful functional-image analysis package. Some calls SPM analysis functions (e.g. GLM) in a batch mode, some were written by myself (e.g. ROI plotting). You probably need some time to learn how to analyze using xjView; but once you learn it, it allows you to analyze your entire data (say 20 subjects) in one day!
link: xjView official website



xjView is being updated right now. The new version will be called xjView 8 and will have the following updates:
1. compatible with SPM8
2. add slice view (or montage view)
3. have an option to render the brain in “old” style
4. single click to have a summary of the current image (including # of clusters, peak MNI coordinate of each cluster, anatomical description of each cluster etc)
xjView 8 is released. http://people.hnl.bcm.tmc.edu/cuixu/xjView/
You can download here
http://www.alivelearn.net/xjview
dear Pro.Cui:
hello,I have a question about Xjview ,this is:
# voxels structure
10 –TOTAL # VOXELS–
10 Left Cerebrum
10 White Matter (L)
6 Frontal Lobe (L)
5 Sub-Gyral (L)
4 Extra-Nuclear (L)
4 Sub-lobar (L)
1 Inferior Frontal Gyrus (L)
how to understand the number?the total of voxels in left cerebrum donot accord with White Matter (L) ,Frontal Lobe (L) , Sub-Gyral (L) , Extra-Nuclear (L), Sub-lobar (L), Inferior Frontal Gyrus (L),why?
look forward to receiving your help,thank a lots
lchy1978
The number means “how many voxels are in xxx region”. The regions listed are not exclusive, meaning a voxel can be both in (e.g.) White Matter and Frontal Lobe. In your case, you have 10 voxels in total; 10 of them are in Left Cerebrum, 10 of them are in White Matter, etc.
@Xu Cui
Dear Xu,
Thank you for these amazing program,
But the link for the manual is not working..
Do you know a way to get the manual.
many thanks
burak
@burak
I will try to put the manual there.
@burak
A manual is put http://www.alivelearn.net/xjview/manual.pdf
Xu, is there a way to choose what slices are shown in the slice view? For example, I want to display only transverse slices -16 to 44?
@Shelli
Shelli, xjview doesn’t offer an explicit option to do that. But it should be fairly easy to do a screen capture and cut extra slices in an image editor (such as photoshop).
Hi,
Our lab recently started using xjview and it has really made our lives easy!I have been having some trouble interpreting the deactivations in xjview and i would be grateful if you can suggest a solution to these:
(1)SPM uses contrast maps (con.img), hence would it be better to use contrast maps itself in xjview? Will using t-maps (spmT.img) make a difference in results?
(2)The deactivation volumes obtained through xjview are not obtained through SPM.Is there a way in which I can calculate the beta values for individual subjects using the deactivation volumes obtained in xjview?
(3) Can the negative z-scores shown in xjview be directly interpreted as deactivations?
@sarika
(1) SPM displays T-test image (not contrast image) in result. They are different. Contrast image saves the difference of beta values, but T image saves the significance (or T value) of the difference.
(2) Assume you have 2 conditions, flash and beep, in your experiment. There will be some regions (e.g. visual cortex) which show greater activity for flash than beep; at the same time, there will be some regions (e.g. auditory cortex) which show greater activity for beep than flash. A contrast of flash-beep will reveal both auditory cortex (positive) and visual cortex (negative). Negative value in visual cortex doesn’t mean “deactivation” of this region, it simply means this region shows less activity in beep than in flash. While SPM only displays one direction (positive direction), spmT images store both information. xjView can show both direction at the same time.
You can’t calculate beta for individual subjects using negative volumes.
(3) No. See point 2 above. btw, it’s usually T-score.
Hi,
I have a basic question. How can I change the color of maps intensity representation with multiple image files? The reason is I want to use montage view, but default is to make the second image colormap yellow, which does not show up very well in publication. When I try to go in and change color properties in Edit->colormap nothing happens.
Best,
Ben
@Ben
inside xjview.m, find function spm_sections, around line 6570, you will find line:
colors = repmat([1 0 0;1 1 0;0 1 0;0 1 1;0 0 1;1 0 1],20,1);
You can change colors of overlay. Each row defines a color in R-G-B value.
e.g. 1 0 0 means red.
To change the corresponding legend color, find function
CallBack_loadImagePush, around line 3411
colours = repmat([1 0 0;1 1 0;0 1 0;0 1 1;0 0 1;1 0 1],100,1);
you should change to same colors.
Hope this helps.
Xu
Thank you for continuing to develop this superb utility. I find it very useful for visualizing and documenting output from many of the standard and experimental analyses that I run (most of which do not involve SPM).
Dear Prof. Xu
I would like to know how to change the first color to blue/green, instead of yellow/red..
I changed the colours when I select 2 t-maps, but I also would like to make pictures with deactivations in blue, instead of red/yellow.
Thanks in advance
clarissa
@clarissa yasuda
Is my answer to Ben above helpful? Here
Dear prof. Cui
I tried these suggestions and I could change the colors of multiple maps. But not for a single T-map. Even with this alterations, the single map display appears in red-yellow..
I would like to use blue-green for displaying deactivations, but I do not know how to change in your script… I am sorry for my ignorance… !!
So, if you have another suggestion…..
Thanks in advance
clarissa
Dear prof. Cui,
I have a double-blind drug-placebo controlled study comparing the difference between groups in response to stress under the influence of drug/ placebo. Stress was administered one time before drug and one time after. SPM gives me values only when there are significant differences. For the second time stress task was administered, there was difference depending on the group type but Not the condition (drug/placebo). SPM indicates there is no significant difference, however, it does not give me any indication of the magnitude of activation in each group. I am asking the following: what does this “insiginificance” mean? It could be that both groups had very high activation magnitude that did not differ between them, or is it that the two groups simply dont have significant activation at all? Thank you so much for your constant help.
@Nattou7
It could be either. Sometimes both groups have nice activation but the “difference” between them is small and still insignificant.
@Xu Cui
Dear Dr. Cui,
How can I know which one of these scenarios is actually the case ? Anyway that SPM would be able to provide me with information about that?
Thank you so much !!
@Malak
When you find A-B is not significant, you will have to do separate contrasts (A-baseline) and (B-baseline) to know which scenario is the case.
What is sub-gyral in xjview and how is it defined? Not familiar with this term:)
@Jacky Rongjun Yu
I have no idea – the anatomical names are from standard database (TD and AAL).
Dear Prof. Xu
I would like to determine the excitation points in human brain ” or initial point of activation in human brain” ????? and how i can determine it in your tool in matlab??
please prof. Xu, can you send to me your email address please ???
Thank you so much !!
@Hossam
Simply click a voxel in the section view and the region name will be displayed.
Dear Prof. Xu
I need your assist in some points i couldn’t reach it:
1. I want some papers talking about excitation points in human brain according to xjView tool, if you have .. can you send me the URLs, if you don’t .. can you tell me the word i must to use in search about these papers, or the method to reach it.
***********************************************************************
2. According to any conditions you determine the excitation points in your brain model??? you choose it random or these points are known??
***********************************************************************
3. I preparing a master in “visualization of the reaction diffusion in brain” .. my work about the propagation of excitation in human brain .. if you anything about this article please tell me
***********************************************************************
please prof.xu assist me .. and can you make your answer in points like i mentioned please
i am really thank you about your previous answer prof. xu, and i hope you give me your email to show you my works if you are interesting ..
[email protected]
@Hossam
1. If you search xjview in google scholar you will find a bunch of papers citing xjview:
http://scholar.google.com/scholar?hl=en&q=xjview&btnG=&as_sdt=1%2C5&as_sdtp=
2. these points are known. I use wfu pickatlas database.
3. Hossam, unfortunately I am not familiar with the field.
xjview 8.10 is out.
Currently it sounds like Movable Type is the best blogging platform available right now.
(from what I’ve read) Is that what you are using on your blog?
Dear Prof. Cui,
I met a trouble and need your assist. After running a conjunction analysis based on SPM8, I view the processed image in Xjview, it appears to give some illogical thresholded p-values. i.e.,when setting p<.5, it always has the message “pValue is too small. No suprathreshold voxels.” This doesn’t make sense. Is there a reason for why this might happen, and/or is there a better way to handle this?
Thanks in advance.
Lifang
Dear Prof. Xu,
I would like to know how to use xjview slice view to select the blobs figure. Where can I get the mannual about the xjview slice view?
Thank you so much !!
@Sugai Liang
What do you mean by ‘select the blobs figure’?
Dear Prof. Xu,
Sorry. Afer the step of results in DARTEL VBM, some blobs can be seen in brain figures. And I would like to know how to use xjview slice view to select the blobs for paper illustration.
Thank you!
xjview 8.13 released
http://www.alivelearn.net/?p=1781
Dear Dr. Cui,
Is there a way for xjview to view .gii (GIFTI) files?
Thank you
@Xu Jing Qian
Unfortunately no.
Dear Dr. Cui
Is there any way to use atlas which is not include in the software, for example:
talairach-daemon-atlas. I have the .nii file from FSL data
Thank you very much.
xiaoxia
@xiaoxia qu
No – but if you can create your own TDdatabase.mat file, then it will work.